Immunotherapies are amazing drugs against cancer. They provide long-lasting benefits, with less side-effects. However, they don’t work all the time. They still fail for too many patients, from 60% to 80% of them. It’s not even known how to correctly identify patients who will benefit from immunotherapies.
In this blog post, I will present two of the best predictors of cancer immunotherapy outcomes, IMPRES and TIDE. I will also show an easy way to combine them and improve their predictions. This will give a new combination biomarker. Finally, I will make suggestions of practical ways to contribute to research on this topic.
You can support my work by making a donation in the donor box here.
You can also ask for collaboration with me on this topic by filling the form.
What is cancer immunotherapy?
Cancer is a disease where various body cells proliferate and become crazy. They run out of control from immune cells, which usually monitor and protect the body.
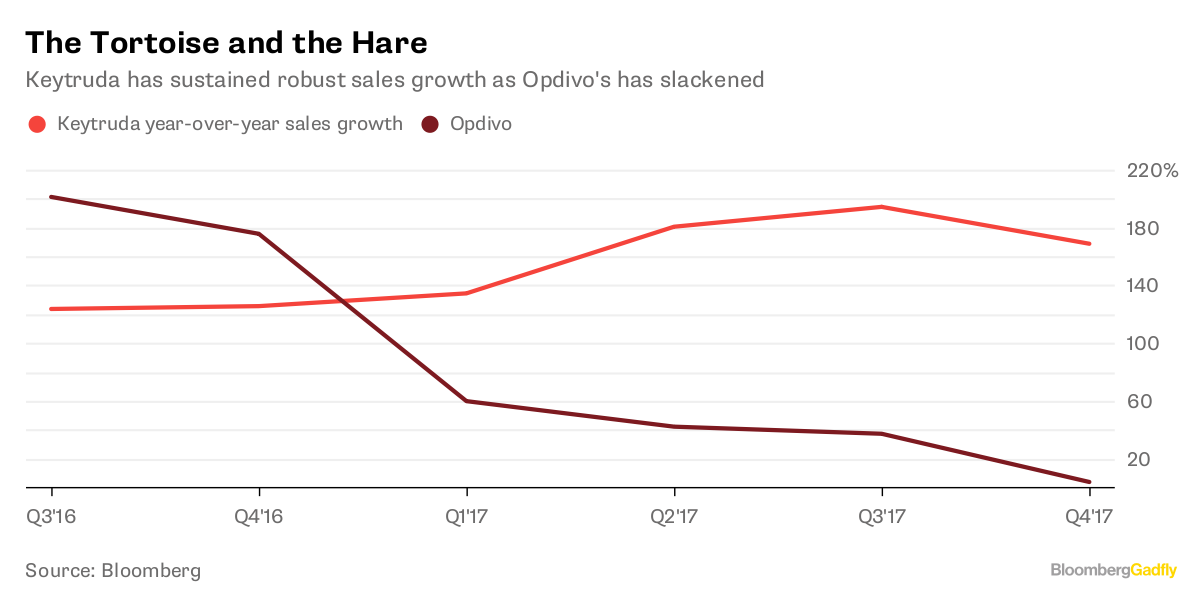
Immunotherapy is a treatment boosting immune cells, to restore their capabilities against cancer. There are different types of immunotherapies, but I focused on Immune Checkpoints Inhibitors, which are now the most popular. These drugs work by ‘inhibiting the inhibitors’ of killer T cells of the immune system. Famous drugs include Opdivo, by Bristol-Myers Squibb, and Keytruda, by Merck & Co. Those two blockbusters had combined sales of almost 14 Billion US dollars in 2018.

To predict immunotherapy results, it’s necessary to look at what’s going on in the tumor. A good way to do that is to measure gene expression of cells.
What is gene expression data?
Most human cells have the same genetic information: 20 000 genes, stored on 23 chromosomes pairs made of DNA.

However, each cell does not express all of those genes uniformly. Gene expression happens when genes, stored on DNA, start producing other molecules called RNA. In turn, RNA is translated into proteins. Proteins are the ‘doers’ of the cell. They perform cell activities.
Gene expression is the number of RNA molecules that a particular gene produces. The type and state of a particular cell are characterized by a pattern of gene expression, called gene signature. Gene expression is measured using a technique called ‘single-cell RNA sequencing’ (scRNA-seq).
At a macroscopic scale, it’s also possible to consider the gene expression of a whole tumor, which is the average of the gene expressions of all the cells inside it. This macroscopic gene expression is measured with a technique called bulk RNA sequencing. It’s a bit older and cheaper than scRNA-seq, because it looks at a more global scale, but it is still used in most cases.

So the data used for predictions is a gene expression matrix, with genes in rows and patients tumors in columns.
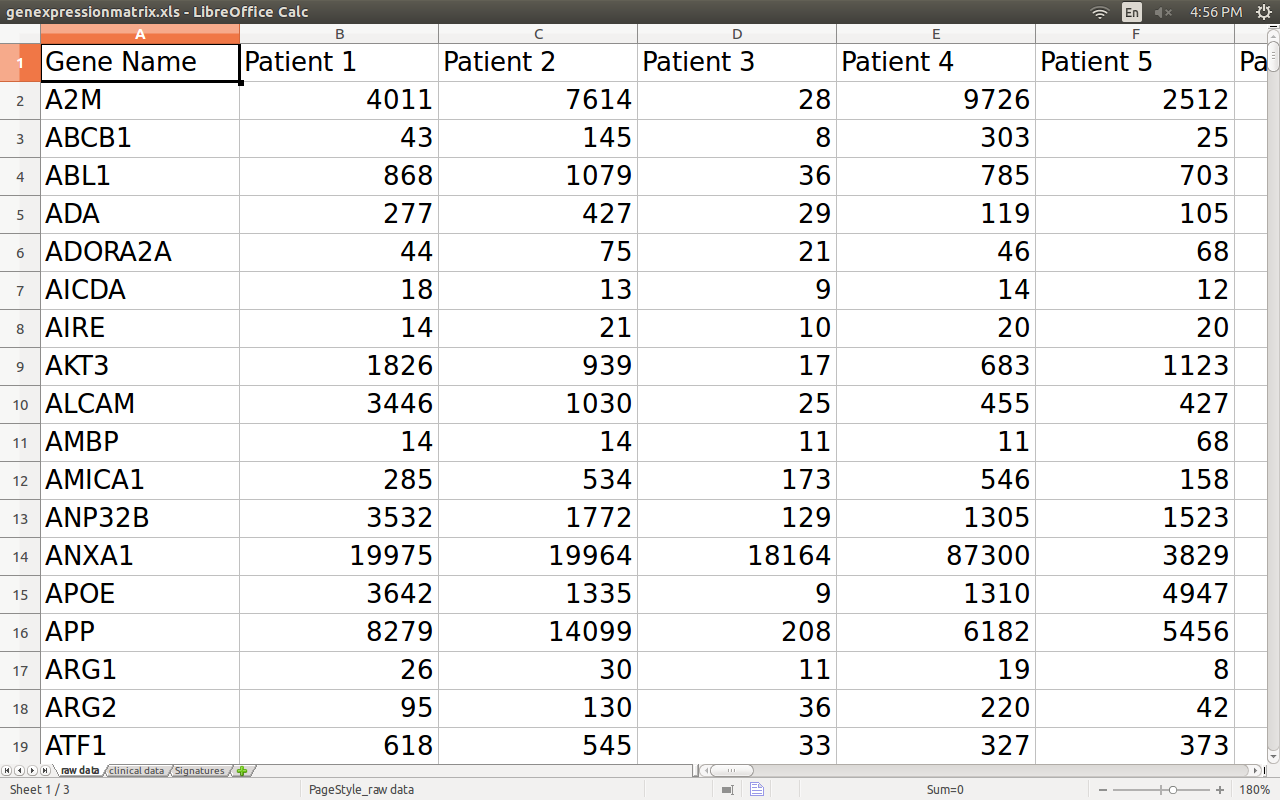
How gene expression data can predict the outcome of cancer immunotherapy
The first predictor is IMPRES, for IMmuno-PREdictive Score (paywalled article, but free with Sci-Hub).
The idea is to look at a particular cancer type called ‘neuroblastoma’. It’s a nerve tissue cancer that usually affects children. In some fortunate cases, the cancer disappears naturally, by the natural action of the immune system.
IMPRES corresponds to a gene signature of those desirable cases. IMPRES is then used as a predictor for the outcome of immunotherapy in all other cancers. It works pretty well: when tested on skin cancer, the IMPRES team reports state-of-the-art performance.
The second predictor is TIDE, for Tumor Immune Dysfunction and Exclusion (paywalled article, but free with Sci-Hub). TIDE is actually the combination of two gene signatures, one for immune dysfunction and another for immune exclusion.
For the dysfunction signature, TIDE identifies genes influencing patient survival in the case when lots of immune T cells infiltrated the tumor (the tumor was “hot”, or “inflammed”). The idea is that if immune cells infiltrated the tumor, but the patient still dies, it means immune cells did not do their job.
The exclusion signature is derived from the gene signatures of three cell types known to restrict T cell infiltration in tumors: cancer-associated fibroblasts, myeloid-derived suppressor cells, and the M2 subtype of tumor-associated macrophages. In this case, the tumor was “cold”, or “excluded”.
30192-9)](https://miro.medium.com/max/375/1*EuNWZbtV1EyRKPL_T_vOlA.jpeg)
Like IMPRES, TIDE also reports state-of-the-art performance. By the way, both papers were simultaneously published in the same journal.
How to make better predictions
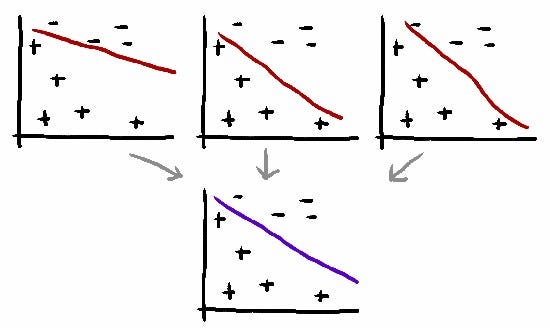
I improved predictions of both IMPRES and TIDE by taking the average of their prediction scores. I got +3.7% improvement over the state-of-the-art.
Technical details are available in my preprint:
Benhenda M. Prediction of clinical response to checkpoint blockade immunotherapy is improved with ensembling. OSF Preprints; 2019. https://doi.org/10.31219/osf.io/r5ke4
Averaging is one of the simplest ensembling method in machine learning. It works because each predictor is focused on different things, so averaging them can cancel out their mistakes.
Impact
My +3.7% improvement is a tiny contribution. However, the overall impact is not negligible, because cancer is a huge global problem. At the current level of use of Keytruda (an immunotherapy with prescription depending on another predictor, TMB), the global impact could be:
- for patients, 2000 human lives/year saved
- for medical insurances (NHS, Medicare…), 100 Million US dollars/year saved
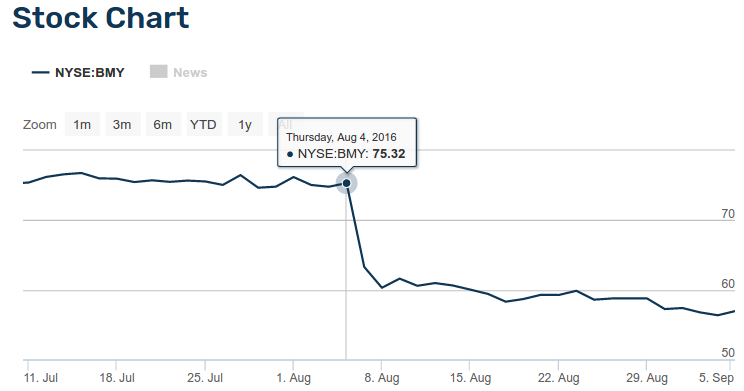
Moreover, pharma and biotech companies need to use the best predictors available, because predictors can make or break FDA approval. In August 5, 2016, the pharma company Bristol-Myers Squibb failed FDA approval for Opdivo in lung cancer, because of improper patient selection for the clinical trial. This failure provoked an immediate reaction of traders in Wall Street, and the company market value suddenly fell by 20 Billion US dollars.
Meanwhile, competitor Merck & Co decided to use a predictor (Tumor Mutation Burden) to select patients for the clinical trial of Keytruda, a drug very-very similar to Opdivo. Keytruda got FDA approval for lung cancer, and it became a success story, with 7 Billion US dollars sales in 2018. A large share of this market would have been taken over by Opdivo, if Bristol-Myers Squibb had implemented better clinical trial design.
How you can contribute
The first possible way to improve predictions is to build the average using the more than 50 predictors currently available. You can also try other ensembling techniques, like stacking.

You can also help finding more data. More data is always more important than better algorithms. Most papers use fewer than 300 patients. That’s 0.3% of the estimated 100 000 patients treated with immune checkpoint inhibitors in 2018 around the world. The remaining 99.7% is buried elsewhere, in private medical records, or is not even collected. Cost of data is not the problem: bulk RNA-sequencing is cheap, $175/sample. It’s less than 0.25% of the cost of immunotherapy, which can reach up to $150 000/year/person in the United States (and at least $70 000/year in other countries).
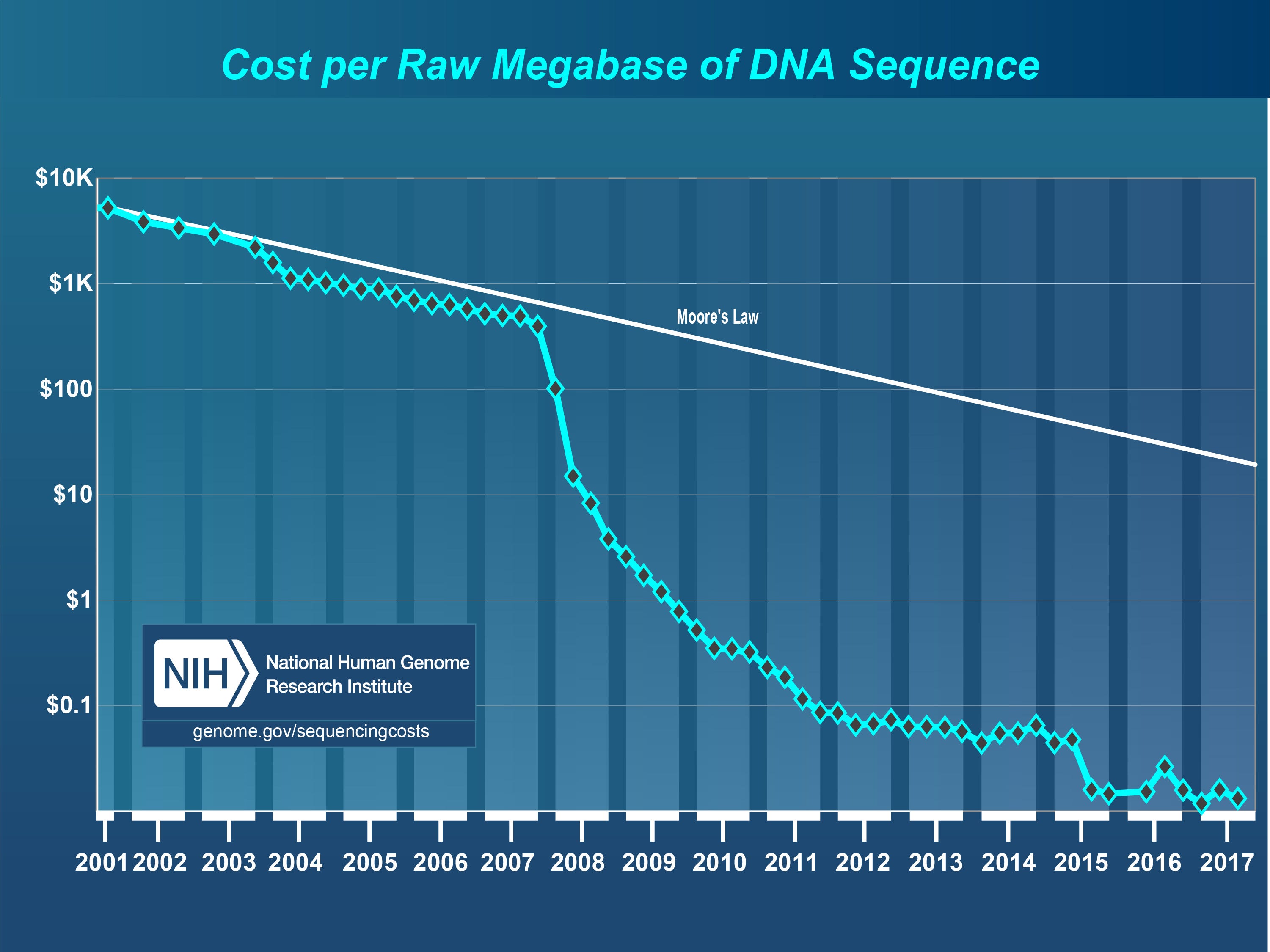
So data scarcity is artificial, and I guess the bottleneck is located in the bureaucracy: med and pharma institutions are big, with a lot of inertia, and they adopt technologies very slowly.

One way to shake bureaucracy is activism. Data activists can accelerate data liberation. Their role is even more important than data scientists, who are constrained by data scarcity. Activists can help bureaucracies realize that cancer lives matter.

Finally, you can contribute by providing incentives and funding to this ecosystem. In particular, I think alternative ways to do research should be encouraged. They can grab low-hanging fruits that remain under the radar of traditional research organizations.

For example, here in my work, I think that ensembling methods were under-appreciated in the immuno-oncology literature before because they were not the best strategy for building Intellectual Property. The averaged predictor is not self-contained, it depends on base predictors that are often proprietary, patented, or otherwise unavailable for commercial use. That’s not the most attractive direction for the industry, or for venture capitalists, who are looking for defensible IP. Likewise, this direction is not favored by academics either, who often use their university labs as launch pads for spin-offs.

In my opinion, that’s the reason why ensembling methods remained a blind spot for the community, even though they are technically simple, and routinely used elsewhere. In conclusion, I think private initiative, and private charity, will remain a critical element of the landscape.
You can support my work by making a donation in the donor box.
You can ask for collaboration with me on this topic by filling the form.
You can also share this blog post with your friends and colleagues.
Initially appeared on Medium.